價格:免費
更新日期:2019-03-22
檔案大小:9.9 MB
目前版本:2.7
版本需求:需要 iOS 12.1 或以上版本。與 iPhone、iPad 及 iPod touch 相容。
支援語言:英語

Isoelectric Point (IEP) is the ultimate tool for accurate estimation of pH value at which the net charge of macromolecule reaches zero.
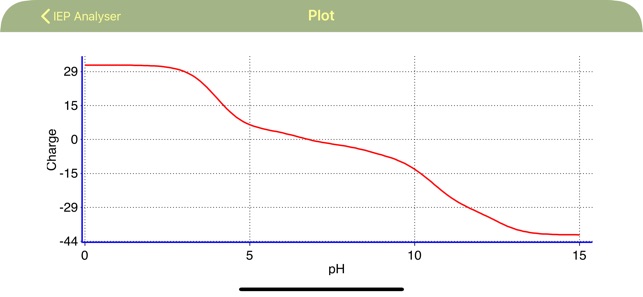
This application calculates relative charge of macromolecule (proteins, peptides etc.), consisting of basic and acidic residues as a function of pH value of solution. The application graphically represents the molecular charge through the whole range of pH values and finds out the isoelectric point value. The application can extract the protein sequence (chain by chain) from FASTA files.
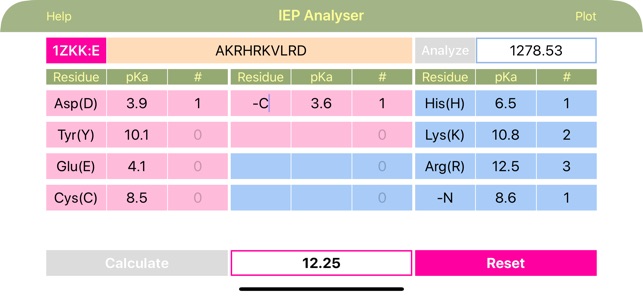
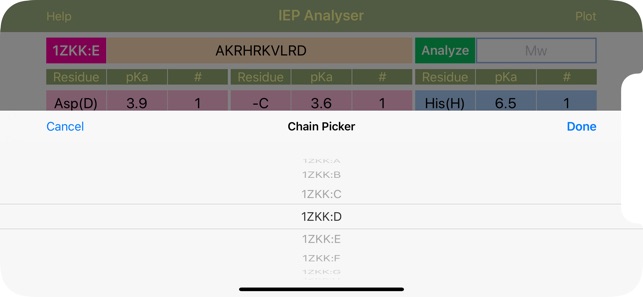
Layout of "IEP". The “Chain” menu allows to choose various chains from the uploaded FASTA file for further analysis. The sequence of polymer molecule is inserted into the “Sequence” field in the single letter format (YHRK…). The sequence can be typed or uploaded from the local FASTA file (for polypeptides). Long press on “Chain” button calls for PDB-styled Fasta-formated text files list located exclusively in “Documents/IEP” folder, from where user can load the file sequence for further analysis. Short press on “Chain” button, calls for chain list (if available) for the current file. The next “Analyze” button, when clicked, analyzes the sequence, calculates the total molar weight and counts and fills the number of recognized acidic and basic residues into the appropriate cells in the amino acid info table. Cells for acidic residues are coloured red and for basic are blue. The table comprised of nine sets of three cells per each amino acid. The three cells provide the charge contributing amino acid acronym, pKa value and number of this particular residue in the macromolecule, respectively. The “-C” and “-N” cells represent standard C- and N-terminus respectively. The “Calculate” button initiates the analysis of the table data (including customized input if made after the sequence analysis) and outputs the IEP value. Important! Table data can be fully customized after or without any sequence analysis! IEP calculation uses table values, whatever their origin.
Application features.
·Estimates the isoelectric point of macromolecule based on pKa values provided.
·Application provides data for most common acidic and basic amino acid residues, but every cell can be customized! Sequence analysis rebuilds the table back to its original state, so customisation should come after initial sequence analysis.
·The application can extract and analyze the protein sequence (chain by chain and all together) from FASTA files.
·The application plots Molar charge vs pH.
Important points.
·“Analyze” button functions: calculation of molar weight and importing sequence data to the table.
·“Calculate” button functions: calculation of IEP based on data in the table.

·If user updates any of the data cells, the program will not start to plot until user press calculate button and only if the update is accepted.
·The cells are colored according to the data input allowed (unchangeable): red for acidic and blue for basic function. However both acidic and basic residue chemical groups are usually provided with pKa values. In case that pKb value is provided, it can be easily converted to pKa using: pKa=pKw-pKb or pKa=14-pKb (at 25°C). User should check what the primary chemical property of the residue is: the proton donor, gaining negative charge (acidic) or proton acceptor, gaining positive charge (basic).
·Unrecognised residue letters are excluded and presented in front of original sequence. The excluded subunits can be added manually to the available cells.
·The empty cells are available for custom input.
·The pKa values can be changed in table to fit the desired model.
·The application assumes no influence between residues.
·The application assigns dot as a decimal separator.
支援平台:iPhone, iPad
